Missing Pieces
About 2 years ago, acwooding and I attended a workshop on text analysis, where a lot of people did some really nice work embedding text into vector spaces under a variety of algorithms. What we were working on was trying to establish some stability results; i.e. whether repeated embeddings the various algorithms were stable, or whether the results were all over the place because, for example, we got results all over the place the algorithm was randomized and we had just gotten lucky.
When we sat down to write up the analysis, we discovered really
quickly that we had a problem. Though we still had a collection of
jupyter notebooks and the associated data blobs, we had no idea how
our collaborators had pre-processed their data to insert into the
process in the first place. We had lost the information about the
preparation of the data, and hence, we’d lost the ability to generate
a consistent set of analyses across all of our data. Our workshop results
weren’t reproducible, and we were going to have to do a bunch of work
over from scratch if we wanted to publish anything.
If you were to survey you average data scientist on how much time they spend in a given phase of the operation, you’d probably get something that looks like this:
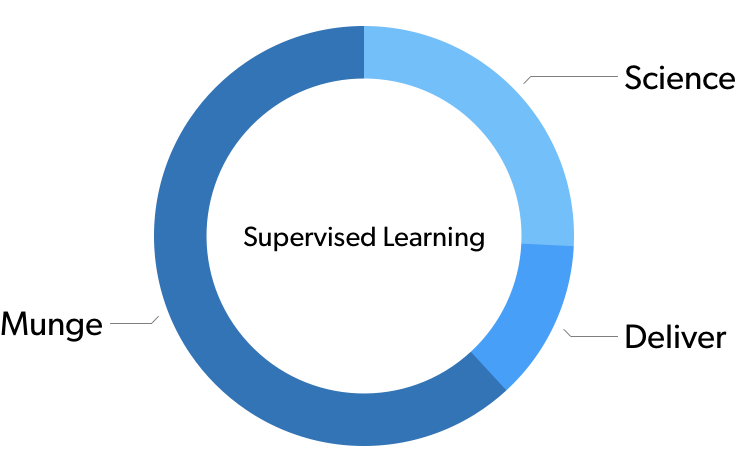
In supervised learning, around 2/3 time is spent munging the data in the first place, before you finally get around to doing your analysis.
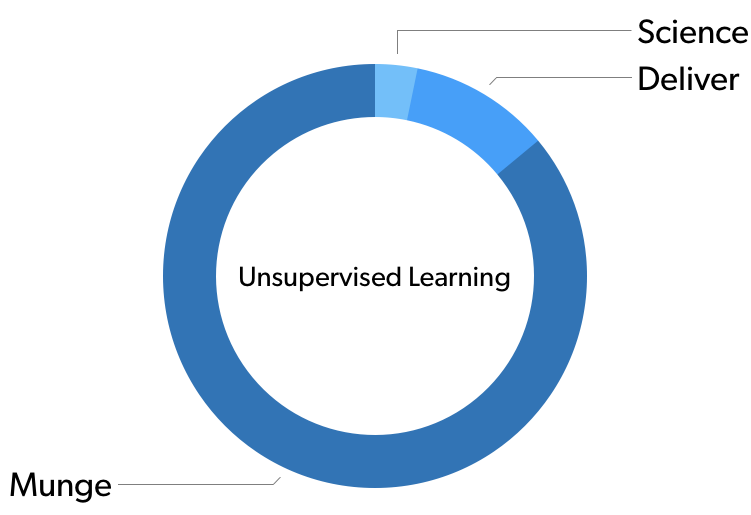
In unsupervised learning problems it’s was more like 90%.
Admittedly, like all statistics, these actual numbers are made up, but they illustrate a real phenomenon. A vast amount of effort we are performing as data scientists is happening before we ever get around to the analysis part.
But What About the Environment?
What we wanted to be able to do was capture that data munging history, and turn that process into something that is sharable and reproducible. We started looking at our own past analyses and set out to create adopt a more standard workflow that would make it easy to preserve (and share) the whole process of data science, including the data munging.
At PyData NYC 2018, we ran a tutorial called “Up your Bus Number: A Reproducible Data Science Workflow.” At that workshop we were intending to talk a great deal about the munging of data, and the wonderful and clever APIs that we had settled on to help simplify that process. When we actually ran the tutorial, it turned out that about 80% of our time was spent before we even got to data munging. it was spent trying to set up consistent, reproducible environments on a wide variety of systems. The hard parts of getting to a reproducible data science pipeline (installing and maintaining your environment), for most of the people we were encountering, didn’t even show up in the survey that we did about where your time is spent. Most people knew of (or even used) tools like anaconda or virtualenv, but not in a way that let them easily maintain and share these environments, or reproduce the environments of others.
Even if we could reproduce the data munging, we couldn’t reproduce the development environment. We have all these fancy tools: anaconda, virtualenv, the now-deceased pipenv, and any number of wrappers and scripts that are designed around making it easier to build a custom python environment that’s tuned for your problem at hand. But actually using those in a consistent manner is not trivial.
When we talk to people about reproducible data science, everyone wants it, but almost nobody wants to do it. Most people think we’re talking about reproducing an analysis, because that’s the easy part. If we dig in a little further, some will grudgingly speak about reproducing their data munging. Almost nobody talks about solving the challenges of reproducible environments, assuming that tools like conda have already solved that. Yet when we sit down to do the work, environment and data munging issues dominate the effort.
Recognizing the Hard Parts
One of our stated goals is to help make data scientists more
productive. How can we do this? Give them the ability to do their job
with less futzing around with their environments, and make it easy for
them to share their work—including the data munging. The primary means
by which data scientists exchange data science lore is by passing
around jupyter notebooks. But there’s so much that goes in under the
hood before that jupyter notebook ever even gets run, that if we don’t
take steps to that additional information—including information about
the environment, the data munging, the metadata associated with the
data sources—then data scientist productivity is lost, and
reproducibility goes right out the window.
 Source: The Turing Way. (CC-BY-4.0)
Source: The Turing Way. (CC-BY-4.0)
Our challenge is this: if we want reproducible data science—and that covers the entire spectrum of reproducibility, replicability, generalizability, and robustness—then the hardest thing we have to do is identify what the hard parts are. The only way to do that is to repeatedly sit down with people and walk through their pipelines. As many people as we can. Take their work and attempt to reproduce it, and in doing so, learn where those barriers to reproduction actually live: the technical barriers, the UX barriers, an the psychology barriers. Then, and only then, put in the hard work of building a toolkit that also solves the psychology and the user interface problems, the workflow, and the APIs associated with preventing reproducibility in the first place.
